
In the central panel, click on the galaxy-gear Convert tab on the top.Click on the galaxy-pencil pencil icon for the dataset to edit its attributes.Six groups are present, with one for each combination of cell type and mouse status. This study examined the expression profiles of basal and luminal cells in the mammary gland of virgin, pregnant and lactating mice. The data for this tutorial comes from a Nature Cell Biology paper, EGF-mediated induction of Mcl-1 at the switch to lactation is essential for alveolar cell survival), Fu et al. We also need some genes to plot in the heatmap.
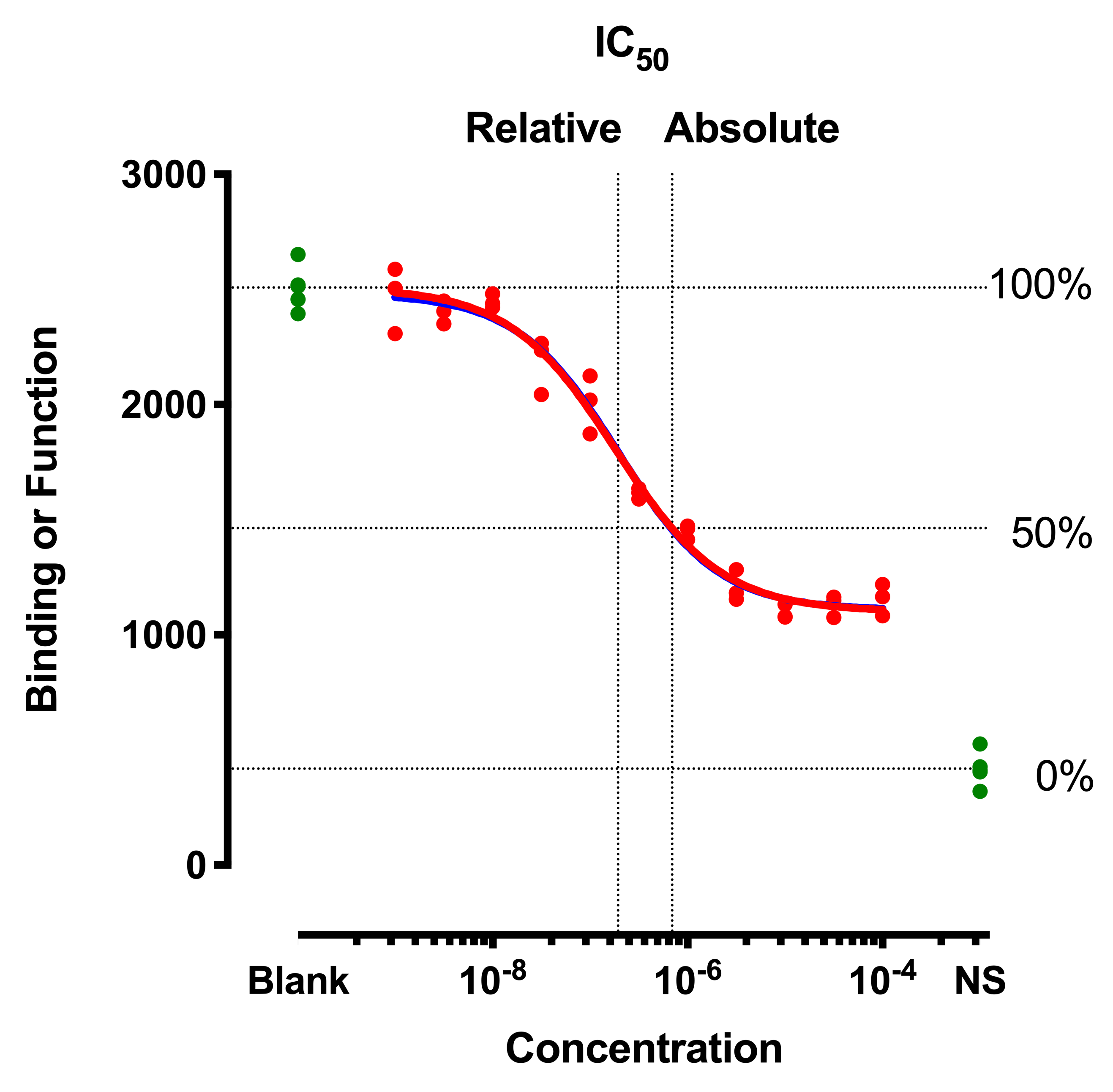
You could also use a file of normalized counts from other RNA-seq differential expression tools, such as edgeR or DESeq2. To generate this file yourself, see the RNA-seq counts to genes tutorial, and run limma-voom selecting “Output Normalised Counts Table?”: Yes. The expression values have been normalized for differences in sequencing depth and composition bias between the samples. To generate a heatmap of RNA-seq results, we need a file of normalized counts.
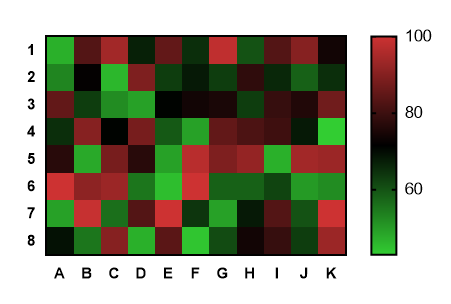
We will also show how a heatmap for a custom set of genes an be created. Here we will demonstrate how to make a heatmap of the top differentially expressed (DE) genes in an RNA-Seq experiment, similar to what is shown for the fruitfly dataset in the RNA-seq ref-based tutorial. The heatmap2 tool uses the heatmap.2 function from the R gplots package. In this tutorial we show how the heatmap2 tool in Galaxy can be used to generate heatmaps.
They are useful for visualizing the expression of genes across the samples. Heatmaps are commonly used to visualize RNA-Seq results.


 0 kommentar(er)
0 kommentar(er)
